Scientific Workflow with HSSM¶

Welcome to the scientific workflow tutorial. This tutorial starts with a basic experimental dataset and we will inch from a very simple HSSM model iteratively toward a model that captures many of the main patterns we can identify in our dataset.
Along the way we try to achieve the following balance:
- Illustrate how HSSM can be used for real scientific workflows. HSSM helps us with model building, running the stats, and reporting results.
- Allow this tutorial to be used as a first look into HSSM, shirking conceptually advanced features that are discused in the many dedicated tutorials you can find on the documentation
Colab Instructions¶
If you would like to run this tutorial on Google colab, please click this link.
Once you are in the colab:
- Follow the installation instructions below (uncomment the respective code)
- restart your runtime.
NOTE:
You may want to switch your runtime to have a GPU or TPU. To do so, go to Runtime > Change runtime type and select the desired hardware accelerator. Note that if you switch your runtime you have to follow the installation instructions again.
Install hssm¶
# If running this on Colab, please uncomment the next line
# !pip install hssm
Download tutorial data¶
# # Data Files
# !wget -P data/carney_workshop_2025_data/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/data/carney_workshop_2025_full.parquet
# !wget -P data/carney_workshop_2025_data/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/data/carney_workshop_2025_modeling.parquet
# !wget -P data/carney_workshop_2025_data/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/data/carney_workshop_2025_parameters.pkl
# # Presampled traces
# !wget -P idata/basic_ddm/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/basic_ddm/traces.nc
# !wget -P idata/ddm_hier/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/ddm_hier/traces.nc
# !wget -P idata/angle_hier/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/angle_hier/traces.nc
# !wget -P idata/angle_hier_v2/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/angle_hier_v2/traces.nc
# !wget -P idata/angle_hier_v3/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/angle_hier_v3/traces.nc
# !wget -P idata/angle_hier_v4/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/angle_hier_v4/traces.nc
# !wget -P idata/angle_v5/ https://raw.githubusercontent.com/lnccbrown/HSSM/main/scientific_workflow_hssm/idata/angle_v5/traces.nc
Start of Tutorial¶
Load modules¶
import hssm
import pandas as pd
import pickle
import numpy as np
import arviz as az
from matplotlib import pyplot as plt
Load workshop data¶
def load_data(filename_base: str,
folder: str = "data") -> tuple[pd.DataFrame, pd.DataFrame, dict]:
"""Load saved simulation data and parameters from files.
Parameters
----------
filename_base : str
Base filename used when saving files
folder : str, optional
Folder containing saved files, by default "data"
Returns
-------
tuple[pd.DataFrame, pd.DataFrame, dict]
Contains:
- DataFrame with modeling data
- DataFrame with full data
- Dict containing group and subject parameters
"""
df_modeling = pd.read_parquet(f"{folder}/{filename_base}_modeling.parquet")
return df_modeling
workshop_data = load_data(filename_base = "carney_workshop_2025",
folder = "scientific_workflow_hssm/data/")
Load Plotting Utilities¶
import matplotlib.pyplot as plt
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
def plot_rt_by_choice(df: pd.DataFrame,
categorical_column: str | None = None,
colors: dict[str, str] | dict[int, str] | None = None,
ax: plt.Axes | None = None):
if categorical_column is None:
ax.hist(df['rt'] * df['response'],
bins = np.linspace(-5,5, 50),
# label=f'Condition {cond}',
histtype='step',
density = True,
color='tab:blue')
else:
for cond in df[categorical_column].unique():
df_cond = df[df[categorical_column] == cond]
ax.hist(df_cond['rt'] * df_cond['response'],
bins = np.linspace(-5,5, 50),
label=f'Condition {cond}',
histtype='step',
density = True,
color=colors[cond])
ax.set_xlabel('RT * Choice')
ax.set_ylabel('Density')
return ax
def inset_bar_plot(df: pd.DataFrame,
categorical_column: str,
response_options: list[int],
colors: dict[str, str] | dict[int, str] | None = None,
ax: plt.Axes | None = None):
axins = inset_axes(ax,
width="35%",
height="35%",
loc='upper left',
borderpad=2.75)
bar_width = 0.55
for j, resp in enumerate(response_options):
for k, cond in enumerate(df[categorical_column].unique()):
k_displace = -1 if k == 0 else 1
df_cond = df[df[categorical_column] == cond]
prop = (df_cond[df_cond.response == resp].shape[0] / len(df_cond))
axins.bar((resp + ((bar_width / 2) * k_displace)),
prop,
width=bar_width,
fill = False,
edgecolor=colors[cond],
label=f'Response {resp}')
axins.set_xticks(response_options)
axins.set_ylim(0, 1)
axins.set_yticks([0.0, 0.5, 1])
axins.set_title('choice proportion / option', fontsize=8)
axins.tick_params(axis='both', which='major', labelsize=7)
axins.set_xlabel('')
axins.set_ylabel('')
return ax
def inset_bar_plot_vertical(df: pd.DataFrame,
categorical_column: str,
response_options: list[int],
colors: dict[str, str] | dict[int, str] | None = None,
ax: plt.Axes | None = None):
axins = inset_axes(ax,
width="35%",
height="35%",
loc='upper left',
borderpad=2.25)
bar_width = 0.55
for j, resp in enumerate(response_options):
# k_displace_dict = {0:}
for k, cond in enumerate(df[categorical_column].unique()):
k_displace = -1 if k == 0 else 1
df_cond = df[df[categorical_column] == cond]
rt_mean = (df_cond[df_cond.response == resp]).rt.mean()
axins.barh((resp + ((bar_width / 2) * k_displace)),
rt_mean,
height=bar_width,
fill = False,
edgecolor=colors[cond],
label=f'Response {resp}')
axins.set_yticks(response_options)
axins.set_xticks([0.0, 1., 2.])
axins.set_title('rt-mean by choice option', fontsize=8)
axins.tick_params(axis='both', which='major', labelsize=7)
axins.set_xlabel('')
axins.set_ylabel('')
return ax
def plot_rt_hists(df: pd.DataFrame,
by_participant: bool = True,
split_by_column: str | None = None,
inset_plot: str | None = "choice proportion",
cols: int = 5):
if split_by_column is not None:
colors = {cond: color for cond, color in zip(df[split_by_column].unique(),
['tab:blue', 'tab:orange', 'tab:green', 'tab:red', 'tab:purple'])}
else:
colors = None
if by_participant:
# Get unique participant IDs and costly_fail_conditions
participants = df['participant_id'].unique()
# Set up subplot grid (adjust cols as needed)
rows = (len(participants) + cols - 1) // cols
fig, axes = plt.subplots(rows, cols,
figsize=(cols*4, rows*3),
sharey=True, sharex=True)
axes = axes.flatten()
for i, pid in enumerate(participants):
ax = axes[i]
df_part = df[df['participant_id'] == pid]
ax = plot_rt_by_choice(df_part,
split_by_column,
colors,
ax)
# Take care of inset plots
if inset_plot == "choice_proportion":
ax = inset_bar_plot(df_part,
split_by_column,
df['response'].unique(),
colors,
ax)
elif inset_plot == "rt_mean":
ax = inset_bar_plot_vertical(df_part,
split_by_column,
df['response'].unique(),
colors,
ax)
if i == 0:
ax.legend(title=split_by_column, loc='best', fontsize='small')
# Hide unused axes
for j in range(i+1, len(axes)):
axes[j].set_visible(False)
plt.tight_layout()
plt.suptitle('RT, Split by Costly Fail Condition and Participant', y=1.02)
plt.show()
else:
fig, ax = plt.subplots(1, 1, figsize=(4, 3))
ax = plot_rt_by_choice(df,
split_by_column,
colors,
ax)
# Take care of inset plots
if inset_plot == "choice_proportion":
ax = inset_bar_plot(df,
split_by_column,
df['response'].unique(),
colors,
ax)
elif inset_plot == "rt_mean":
ax = inset_bar_plot_vertical(df,
split_by_column,
df['response'].unique(),
colors,
ax)
ax.legend(title=split_by_column, loc='best', fontsize='small')
plt.tight_layout()
plt.suptitle('RT by Trial, Split by Costly Fail Condition', y=1.02)
plt.show()
Exploratory Data Analysis¶

Now that we are done preparing the setup, let's get to the meat of it! The picture above gives us a bit of an idea, where the dataset that we are going to work with below comes from (alert: the backstory may or may not be real).
20 subjects, performed 250 trials each of a basic Random dot motion task. The task seemingly had two important manipulations.
- A costly fail condition, in which subjects get punished for mistakes.
- A trial by trail manipulation of difficulty (in the Random dot motion task, this refers to degree of coherence with which the dots move in a particular direction)
Let's take a look at the actual dataframe.
workshop_data
| response | rt | participant_id | trial | costly_fail_condition | continuous_difficulty | response_l1 | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.556439 | 0 | 1 | 1 | -0.277337 | 0 |
| 1 | 1 | 0.741682 | 0 | 2 | 0 | -0.810919 | 1 |
| 2 | 1 | 0.461832 | 0 | 3 | 0 | -0.673330 | 1 |
| 3 | 1 | 0.626154 | 0 | 4 | 0 | 0.755445 | 1 |
| 4 | 1 | 0.651677 | 0 | 5 | 1 | 0.136755 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 4995 | 1 | 1.039342 | 19 | 246 | 0 | -0.612223 | -1 |
| 4996 | 1 | 1.587827 | 19 | 247 | 0 | 0.732396 | 1 |
| 4997 | 1 | 0.668594 | 19 | 248 | 1 | -0.175321 | 1 |
| 4998 | 1 | 1.616471 | 19 | 249 | 0 | -0.630447 | 1 |
| 4999 | 1 | 1.051329 | 19 | 250 | 1 | 0.511197 | 1 |
5000 rows × 7 columns
Adding a few columns¶
As part of prep work for plotting etc. we will add a few columns here. These will be motivated later (close your eyes :)).
# Binary version of difficulty
workshop_data['bin_difficulty'] = workshop_data['continuous_difficulty'].apply(lambda x: 'high' if x > 0 else 'low')
# I want a a ordinal variable that is composed of 5 quantile levels of difficulty
workshop_data['quantile_difficulty'] = pd.qcut(workshop_data['continuous_difficulty'],
3, labels = ['-1', '0', '1'])
workshop_data['quantile_difficulty_binary'] = pd.qcut(workshop_data['continuous_difficulty'],
2, labels = ['-1', '1'])
# Slightly
workshop_data['response_l1_plotting'] = workshop_data['response_l1'].apply(lambda x: str(-1) if x == -1 else str(1))
Most basic reaction time plot¶
plot_rt_hists(workshop_data,
by_participant = False,
split_by_column = None,
inset_plot = None)
/var/folders/gx/s43vynx550qbypcxm83fv56dzq4hgg/T/ipykernel_64548/900136031.py:166: UserWarning: No artists with labels found to put in legend. Note that artists whose label start with an underscore are ignored when legend() is called with no argument. ax.legend(title=split_by_column, loc='best', fontsize='small')
So far so good. Looking at the global reaction time pattern, it does seem commensurate with what we might expect out of basic Sequential Sampling Model (SSM). The basic DDM might be a good start here.
Basic Model: DDM¶
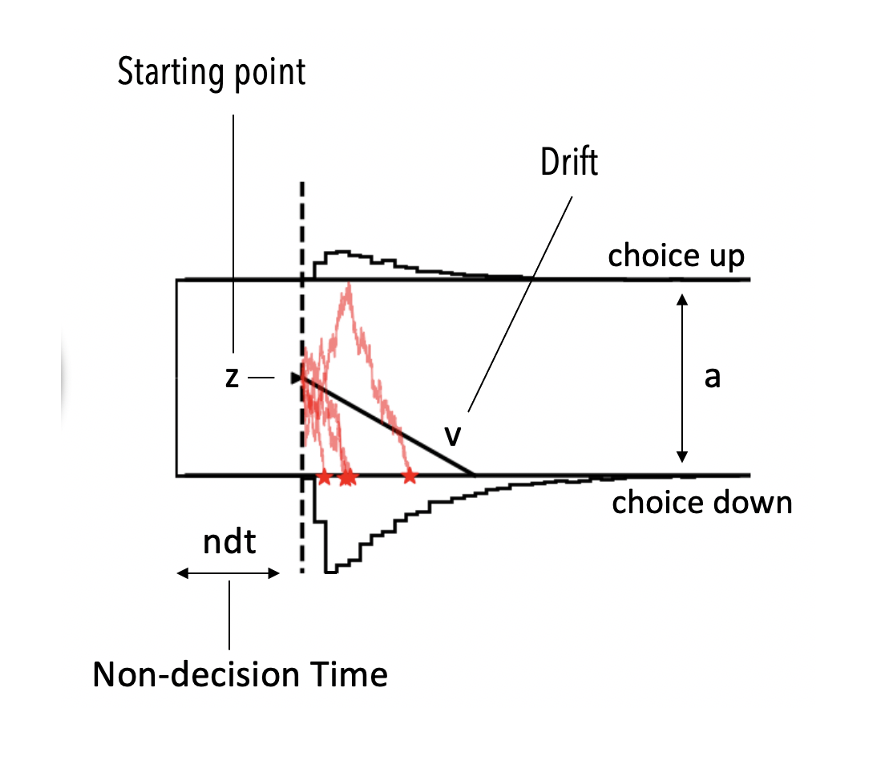
The picture above illustrates the basic Drift Diffusion Model. Note the parameters,
vthe drift rate (how much evidence do I collect on average per unit of time)athe boundary separation (how much evidence do I need to commit to a choice)zhow biased am I toward a particular choice a priorindt(we will simply call ittbelow), the delay between being exposed to a stimulus and starting the actual evidence accumulation process
BasicDDMModel = hssm.HSSM(data = workshop_data,
model = "ddm",
loglik_kind = "approx_differentiable",
global_formula = "y ~ 1",
noncentered = False,
)
Model initialized successfully.
BasicDDMModel
Hierarchical Sequential Sampling Model
Model: ddm
Response variable: rt,response
Likelihood: approx_differentiable
Observations: 5000
Parameters:
v:
Formula: v ~ 1
Priors:
v_Intercept ~ Normal(mu: 0.0, sigma: 0.25)
Link: identity
Explicit bounds: (-3.0, 3.0)
a:
Formula: a ~ 1
Priors:
a_Intercept ~ Normal(mu: 1.4, sigma: 0.25)
Link: identity
Explicit bounds: (0.3, 2.5)
z:
Formula: z ~ 1
Priors:
z_Intercept ~ Normal(mu: 0.5, sigma: 0.25)
Link: identity
Explicit bounds: (0.0, 1.0)
t:
Formula: t ~ 1
Priors:
t_Intercept ~ Normal(mu: 1.0, sigma: 0.25)
Link: identity
Explicit bounds: (0.0, 2.0)
Lapse probability: 0.05
Lapse distribution: Uniform(lower: 0.0, upper: 20.0)
BasicDDMModel.graph()
try:
# Load pre-computed traces
BasicDDMModel.restore_traces(traces = "scientific_workflow_hssm/idata/basic_ddm/traces.nc")
except:
# Sample posterior
basic_ddm_idata = BasicDDMModel.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
BasicDDMModel.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
BasicDDMModel.save_model(model_name = "basic_ddm",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
BasicDDMModel.traces
-
<xarray.Dataset> Size: 36kB Dimensions: (chain: 2, draw: 500) Coordinates: * chain (chain) int64 16B 0 1 * draw (draw) int64 4kB 0 1 2 3 4 5 6 ... 493 494 495 496 497 498 499 Data variables: t_Intercept (chain, draw) float64 8kB ... z_Intercept (chain, draw) float64 8kB ... a_Intercept (chain, draw) float64 8kB ... v_Intercept (chain, draw) float64 8kB ... Attributes: created_at: 2025-07-11T17:29:22.668745+00:00 arviz_version: 0.21.0 inference_library: numpyro inference_library_version: 0.17.0 sampling_time: 95.06823 tuning_steps: 500 modeling_interface: bambi modeling_interface_version: 0.15.0 -
<xarray.Dataset> Size: 32MB Dimensions: (chain: 2, draw: 200, __obs__: 5000, rt,response_dim: 2) Coordinates: * chain (chain) int64 16B 0 1 * draw (draw) int64 2kB 0 1 2 3 4 5 6 ... 194 195 196 197 198 199 * __obs__ (__obs__) int64 40kB 0 1 2 3 4 ... 4995 4996 4997 4998 4999 * rt,response_dim (rt,response_dim) int64 16B 0 1 Data variables: rt,response (chain, draw, __obs__, rt,response_dim) float64 32MB ... Attributes: modeling_interface: bambi modeling_interface_version: 0.15.0 -
<xarray.Dataset> Size: 40MB Dimensions: (chain: 2, draw: 500, __obs__: 5000) Coordinates: * chain (chain) int64 16B 0 1 * draw (draw) int64 4kB 0 1 2 3 4 5 6 ... 493 494 495 496 497 498 499 * __obs__ (__obs__) int64 40kB 0 1 2 3 4 5 ... 4995 4996 4997 4998 4999 Data variables: rt,response (chain, draw, __obs__) float64 40MB ... Attributes: modeling_interface: bambi modeling_interface_version: 0.15.0 -
<xarray.Dataset> Size: 53kB Dimensions: (chain: 2, draw: 500) Coordinates: * chain (chain) int64 16B 0 1 * draw (draw) int64 4kB 0 1 2 3 4 5 6 ... 494 495 496 497 498 499 Data variables: acceptance_rate (chain, draw) float64 8kB ... step_size (chain, draw) float64 8kB ... diverging (chain, draw) bool 1kB ... energy (chain, draw) float64 8kB ... n_steps (chain, draw) int64 8kB ... tree_depth (chain, draw) int64 8kB ... lp (chain, draw) float64 8kB ... Attributes: created_at: 2025-07-11T17:29:22.675898+00:00 arviz_version: 0.21.0 modeling_interface: bambi modeling_interface_version: 0.15.0 -
<xarray.Dataset> Size: 120kB Dimensions: (__obs__: 5000, rt,response_extra_dim_0: 2) Coordinates: * __obs__ (__obs__) int64 40kB 0 1 2 3 ... 4997 4998 4999 * rt,response_extra_dim_0 (rt,response_extra_dim_0) int64 16B 0 1 Data variables: rt,response (__obs__, rt,response_extra_dim_0) float64 80kB ... Attributes: created_at: 2025-07-11T17:29:22.676800+00:00 arviz_version: 0.21.0 inference_library: numpyro inference_library_version: 0.17.0 sampling_time: 95.06823 tuning_steps: 500 modeling_interface: bambi modeling_interface_version: 0.15.0
az.summary(BasicDDMModel.traces)
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| t_Intercept | 0.328 | 0.005 | 0.317 | 0.338 | 0.000 | 0.000 | 576.0 | 573.0 | 1.0 |
| z_Intercept | 0.466 | 0.007 | 0.453 | 0.478 | 0.000 | 0.000 | 529.0 | 496.0 | 1.0 |
| a_Intercept | 1.017 | 0.009 | 1.000 | 1.033 | 0.000 | 0.000 | 583.0 | 610.0 | 1.0 |
| v_Intercept | 0.943 | 0.023 | 0.900 | 0.985 | 0.001 | 0.001 | 536.0 | 526.0 | 1.0 |
az.plot_trace(BasicDDMModel.traces)
plt.tight_layout()
az.plot_forest(BasicDDMModel.traces)
plt.tight_layout()
az.plot_pair(BasicDDMModel.traces,
kind="kde",
marginals=True)
array([[<Axes: ylabel='t_Intercept'>, <Axes: >, <Axes: >, <Axes: >],
[<Axes: ylabel='z_Intercept'>, <Axes: >, <Axes: >, <Axes: >],
[<Axes: ylabel='a_Intercept'>, <Axes: >, <Axes: >, <Axes: >],
[<Axes: xlabel='t_Intercept', ylabel='v_Intercept'>,
<Axes: xlabel='z_Intercept'>, <Axes: xlabel='a_Intercept'>,
<Axes: xlabel='v_Intercept'>]], dtype=object)
ax = hssm.plotting.plot_model_cartoon(
BasicDDMModel,
n_samples=10,
bins=20,
plot_pp_mean=True,
plot_pp_samples=False,
n_trajectories=2, # extra arguments for the underlying plot_model_cartoon() function
);
No posterior predictive samples found. Generating posterior predictive samples using the provided InferenceData object and the original data. This will modify the provided InferenceData object, or if not provided, the traces object stored inside the model.
Output()
Output()
Output()
Output()
ax = hssm.plotting.plot_quantile_probability(BasicDDMModel,
cond="quantile_difficulty",
)
ax.set_ylim(0, 3);
# ax.set_xlim(-0.1, 1.1);
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:327: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond, "is_correct"])["rt"] /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:334: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond])["is_correct"]
ax = hssm.plotting.plot_quantile_probability(BasicDDMModel,
cond="costly_fail_condition",
)
ax.set_ylim(0, 3);
ax = hssm.plotting.plot_quantile_probability(BasicDDMModel,
cond="response_l1_plotting",
)
ax.set_ylim(0, 3);
# Posterior predictive
BasicDDMModel.plot_predictive(step = True,
col = 'participant_id',
col_wrap = 5,
bins = np.linspace(-5,5, 50))
<seaborn.axisgrid.FacetGrid at 0x2ba7a3110>
Taking stock¶
We can observe a few patterns here.
- First, cleary the reaction time distributions are not the same for every subject, we need to account for that.
- Second, I does seem like the tail of the reaction time distribution is more graceful in for our predictions than it is in the original subject data. (This was less clear when looking only at the global pattern...)
We will now adjust our model to tackle these patterns one by one. Let's begin by specializing our parameters by subject.
In Bayesian Inference we approach this by introducing a Hierarchy, we assume that subject level parameters derive from a common group distribution.
Inference then proceeds over the parameters of this group distribution, as well as the subject wise parameters.
Hierarchies serve as a form of regularization of our parameter estimates, the group distribution allows us to share information between the single subject parameters estimates.
You don't have to use a hierarchy, we could introduce a subject wise parameterization e.g. by simply treating participant_id as a categorical variable / collection of dummy variables without using any notion of a group distribution (and you are welcome to try this).
DDM Hierarchical¶
Moving on to our first hierarchical model. As a first step, we will use our global_formula argument to (1|participant_id), which is equivalent to 1 + (1|participant_id),
(use 0 + (1|participant_id) is you explicitly don't want to create an intercept).
This will make all parameters of our model hierarchical.
DDMHierModel = hssm.HSSM(data = workshop_data,
model = "ddm",
loglik_kind = "approx_differentiable",
global_formula = "y ~ (1|participant_id)", # New
noncentered = False,
)
Model initialized successfully.
try:
# Load pre-computed traces
DDMHierModel.restore_traces(traces = "scientific_workflow_hssm/idata/ddm_hier/traces.nc")
except:
# Sample posterior
ddm_hier_idata = DDMHierModel.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
DDMHierModel.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
DDMHierModel.save_model(model_name = "ddm_hier",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
DDMHierModel.graph()
az.plot_trace(DDMHierModel.traces)
plt.tight_layout()
ax = hssm.plotting.plot_model_cartoon(
DDMHierModel,
col = "participant_id",
col_wrap = 5,
n_samples=100,
bin_size=0.2,
plot_pp_mean=True,
# color_pp_mean = "red",
# color_pp = "black",
plot_pp_samples=False,
n_trajectories=2, # extra arguments for the underlying plot_model_cartoon() function
);
No posterior predictive samples found. Generating posterior predictive samples using the provided InferenceData object and the original data. This will modify the provided InferenceData object, or if not provided, the traces object stored inside the model.
Output()
Output()
Output()
Output()
Comparing Parameter Loadings¶
az.summary(DDMHierModel.traces,
filter_vars = "like",
var_names = ["~participant_id"]).sort_index()
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| a_Intercept | 1.041 | 0.019 | 1.005 | 1.078 | 0.001 | 0.001 | 311.0 | 398.0 | 1.01 |
| t_Intercept | 0.375 | 0.021 | 0.337 | 0.414 | 0.002 | 0.002 | 89.0 | 121.0 | 1.01 |
| v_Intercept | 0.948 | 0.110 | 0.734 | 1.146 | 0.010 | 0.006 | 121.0 | 205.0 | 1.01 |
| z_Intercept | 0.455 | 0.016 | 0.424 | 0.485 | 0.001 | 0.001 | 190.0 | 259.0 | 1.01 |
az.summary(BasicDDMModel.traces).sort_index()
| mean | sd | hdi_3% | hdi_97% | mcse_mean | mcse_sd | ess_bulk | ess_tail | r_hat | |
|---|---|---|---|---|---|---|---|---|---|
| a_Intercept | 1.017 | 0.009 | 1.000 | 1.033 | 0.000 | 0.000 | 583.0 | 610.0 | 1.0 |
| t_Intercept | 0.328 | 0.005 | 0.317 | 0.338 | 0.000 | 0.000 | 576.0 | 573.0 | 1.0 |
| v_Intercept | 0.943 | 0.023 | 0.900 | 0.985 | 0.001 | 0.001 | 536.0 | 526.0 | 1.0 |
| z_Intercept | 0.466 | 0.007 | 0.453 | 0.478 | 0.000 | 0.000 | 529.0 | 496.0 | 1.0 |
The mean parameters of our models are de facto quite similar. Allowing subject wise variation however dramatically improved our fit to the data!
Quantitative Model Comparison¶
az.compare(
{"DDM": BasicDDMModel.traces,
"DDM Hierarchical": DDMHierModel.traces}
)
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn(
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| DDM Hierarchical | 0 | -4954.407753 | 74.496783 | 0.000000 | 0.999617 | 76.438410 | 0.000000 | True | log |
| DDM | 1 | -5645.531361 | 4.771162 | 691.123609 | 0.000383 | 74.728599 | 32.604471 | False | log |
Comparing predictions¶
# Posterior predictive
DDMHierModel.plot_predictive(step = True,
col_wrap = 5,
bins = np.linspace(-5,5, 50));
# Posterior predictive
BasicDDMModel.plot_predictive(step = True,
col_wrap = 5,
bins = np.linspace(-5,5, 50));
# Posterior predictive
DDMHierModel.plot_predictive(step = True,
col = 'participant_id',
col_wrap = 5,
bins = np.linspace(-5,5, 50))
<seaborn.axisgrid.FacetGrid at 0x2d129f850>
Taking Stock¶
Let's take stock again of any obvious pontential for improving our model here. We are now capturing the data much better subject by subject, however looking closely,
it seems like the tail behavior of the observed and the predicted data is somewhat different, for a few subjects.
The particularly suspicious subjest, are:
participant_id = 1participant_id = 14participant_id = 15participant_id = 17
It seems that for these (and there are others) participants, the model predicted data has a wider tail than what we actually observe in our dataset.
This will motivate a change in the Sequential Sampling Model that we apply.
Angle Model Hierarchical¶
Given what we concluded about the tail behavior of the observed RTs, we will adjust our SSM, to allow for linear collapsing bounds. HSSM ships with a such a model,
and we can apply it to our data simple by changing the model argument. The corresponding model is called angle model in our lingo, and is illustrated below conceptually.
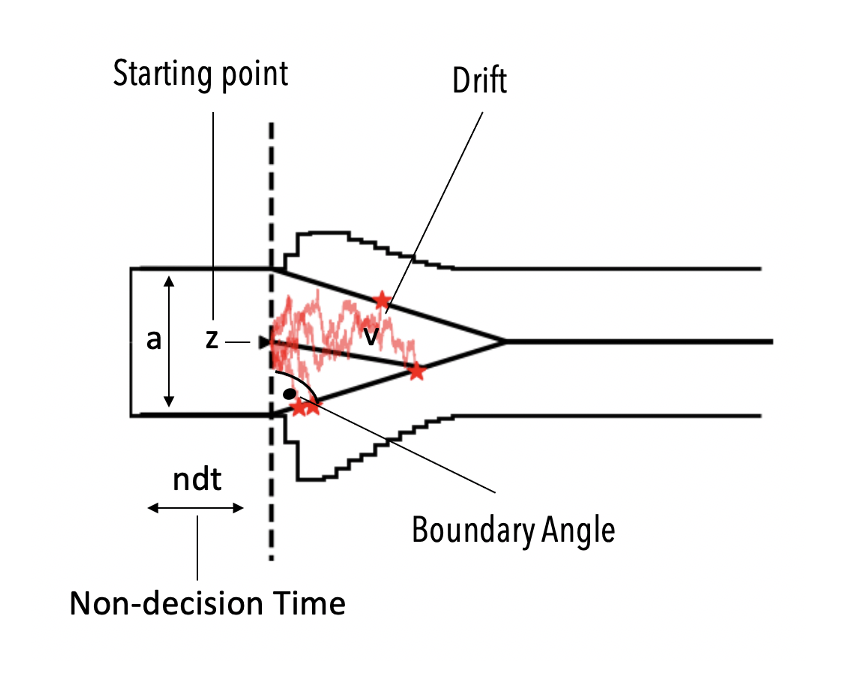
AngleHierModel = hssm.HSSM(data = workshop_data,
model = "angle",
loglik_kind = "approx_differentiable",
global_formula = "y ~ (1|participant_id)",
noncentered = False,
)
Model initialized successfully.
AngleHierModel.graph()
try:
# Load pre-computed traces
AngleHierModel.restore_traces(traces = "scientific_workflow_hssm/idata/angle_hier/traces.nc")
except:
# Sample posterior
angle_hier_idata = AngleHierModel.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
AngleHierModel.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
AngleHierModel.save_model(model_name = "angle_hier",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
ax = hssm.plotting.plot_model_cartoon(
AngleHierModel,
col = 'participant_id',
col_wrap = 5,
n_samples=10,
bin_size=0.2,
plot_pp_mean=True,
plot_pp_samples=False,
n_trajectories=2, # extra arguments for the underlying plot_model_cartoon() function
);
No posterior predictive samples found. Generating posterior predictive samples using the provided InferenceData object and the original data. This will modify the provided InferenceData object, or if not provided, the traces object stored inside the model.
Output()
Output()
Output()
Output()
Output()
We can up it one notch and include the parameter uncertainty in the model_cartoon_plot(). This helps us assess how certain we are about the setting of the boundary collapse here.
Let's see what that looks like!
ax = hssm.plotting.plot_model_cartoon(
AngleHierModel,
col = 'participant_id',
col_wrap = 5,
n_samples=50,
bin_size=0.2,
plot_pp_mean=True,
plot_pp_samples=True,
n_trajectories=2, # extra arguments for the underlying plot_model_cartoon() function
);
No posterior predictive samples found. Generating posterior predictive samples using the provided InferenceData object and the original data. This will modify the provided InferenceData object, or if not provided, the traces object stored inside the model.
Output()
Output()
Output()
Output()
Output()
Output()
Output()
Output()
Output()
Output()
Angle (theta) parameter Bayesian t-test¶
az.plot_posterior(AngleHierModel.traces,
var_names = ["theta_Intercept"],
ref_val = 0,
kind = "hist",
ref_val_color = "red",
histtype = "step")
<Axes: title={'center': 'theta_Intercept'}>
# Posterior predictive
AngleHierModel.plot_predictive(step = True,
col_wrap = 5,
bins = np.linspace(-5,5, 50))
<Axes: title={'center': 'Posterior Predictive Distribution'}, xlabel='Response Time', ylabel='Density'>
# Posterior predictive
AngleHierModel.plot_predictive(step = True,
col = 'participant_id',
col_wrap = 5,
bins = np.linspace(-5,5, 50))
<seaborn.axisgrid.FacetGrid at 0x3124ee5d0>
Visually it seems like we did improve the fit (even though the difference in visual improvement is much less than what we had witnessed introducing the hierarchy in the first place).
Let us corroborate the visual intuition via formal model comparison.
Quantitative Model Comparison¶
az.compare(
{"DDM": BasicDDMModel.traces,
"DDM Hierarchical": DDMHierModel.traces,
"Angle Hierarchical": AngleHierModel.traces}
)
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn(
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| Angle Hierarchical | 0 | -4736.150740 | 78.295641 | 0.000000 | 1.000000e+00 | 72.823473 | 0.000000 | False | log |
| DDM Hierarchical | 1 | -4954.407753 | 74.496783 | 218.257012 | 1.547572e-07 | 76.438410 | 16.444052 | True | log |
| DDM | 2 | -5645.531361 | 4.771162 | 909.380621 | 0.000000e+00 | 74.728599 | 36.362620 | False | log |
Good, introducing the angle model seemed to have helped us quite a bit, even though, as intuited by the simple visual inspection, the improvement in elpd_loo is not as
the improvement in going from a simple model toward a hierarchical model (even though the actual SSM was misspecified).
So what next? On the surface, it looks like we have a model that fits our data quite well.
Let's take another look at our data to identify more patterns that we may not capture with out current efforts.
Further EDA¶
Maybe it is time to look more directly at the effects of our experiment manipulations.
Below are a few graphs to understand what might be happening.
# Posterior predictive
AngleHierModel.plot_predictive(step = True,
col = 'costly_fail_condition',
bins = np.linspace(-5,5, 50))
plt.tight_layout()
ax = hssm.plotting.plot_quantile_probability(AngleHierModel,
cond="costly_fail_condition",
)
ax.set_ylim(0, 3);
plot_rt_hists(workshop_data,
by_participant = True,
split_by_column = "costly_fail_condition",
inset_plot = "choice_proportion")
/var/folders/gx/s43vynx550qbypcxm83fv56dzq4hgg/T/ipykernel_64548/900136031.py:142: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect. plt.tight_layout()
plot_rt_hists(workshop_data,
by_participant = True,
split_by_column = "costly_fail_condition",
inset_plot = "rt_mean")
/var/folders/gx/s43vynx550qbypcxm83fv56dzq4hgg/T/ipykernel_64548/900136031.py:142: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect. plt.tight_layout()
We can identify two patterns.
- On average in the
costly_fail_conditionparticipants seem to make slightly fewer mistakes - On average in the
costly_fail_conditionparticipants seem to take a little longer for their choices!
This meshes with how we expect the incentives to act. Participants should be slightly more cautious to get it right, if mistakes are costly!
In the contect of SSMs, this is usually mapped on to the decision threshold (parameter a), so maybe we should try to incorporate the costly_fail_condition in the regression
function for that parameter in our model.
Addressing costly fail condition¶
To include parameter specific regressions, we can rely on the include argument in HSSM. Let's illustrate this.
AngleHierModelV2 = hssm.HSSM(data = workshop_data,
model = "angle",
loglik_kind = "approx_differentiable",
global_formula = "y ~ (1|participant_id)",
include = [{"name": "a",
"formula": "a ~ (1 + C(costly_fail_condition)|participant_id)"}],
noncentered = False,
)
Model initialized successfully.
AngleHierModelV2.graph()
try:
# Load pre-computed traces
AngleHierModelV2.restore_traces(traces = "scientific_workflow_hssm/idata/angle_hier_v2/traces.nc")
except:
# Sample posterior
angle_hier_idata = AngleHierModelV2.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
AngleHierModelV2.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
AngleHierModelV2.save_model(model_name = "angle_hier_v2",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
az.plot_trace(AngleHierModelV2.traces)
plt.tight_layout()
az.plot_posterior(AngleHierModelV2.traces,
var_names = ["a_C(costly_fail_condition)|participant_id_mu"],
ref_val = 0,
kind = "hist",
ref_val_color = "red",
histtype = "step")
<Axes: title={'center': 'a_C(costly_fail_condition)|participant_id_mu\n1'}>
# Posterior predictive
AngleHierModelV2.plot_predictive(step = True,
# row = 'participant_id',
col = 'costly_fail_condition',
bins = np.linspace(-5, 5, 50),
)
plt.tight_layout()
plt.show()
ax = hssm.plotting.plot_quantile_probability(AngleHierModelV2,
cond="costly_fail_condition",
)
ax.set_ylim(0, 3);
Quite an improvement! Let's see what our quantitative model comparison metrics say.
Quantitative Model Comparison¶
az.compare(
{"DDM": BasicDDMModel.traces,
"DDM Hierarchical": DDMHierModel.traces,
"Angle Hierarchical": AngleHierModel.traces,
"Angle Hierarchical Cost": AngleHierModelV2.traces}
)
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn(
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| Angle Hierarchical Cost | 0 | -4470.826272 | 87.516854 | 0.000000 | 1.000000e+00 | 70.898643 | 0.000000 | False | log |
| Angle Hierarchical | 1 | -4736.150740 | 78.295641 | 265.324468 | 2.313717e-07 | 72.823473 | 20.493430 | False | log |
| DDM Hierarchical | 2 | -4954.407753 | 74.496783 | 483.581480 | 1.523141e-07 | 76.438410 | 25.173534 | True | log |
| DDM | 3 | -5645.531361 | 4.771162 | 1174.705089 | 0.000000e+00 | 74.728599 | 40.610196 | False | log |
Good, we now incorporated the costly_fail_condition in a conceptually coherent manner.
Let's take a look at difficulty next.
ax = hssm.plotting.plot_quantile_probability(AngleHierModelV2,
cond="quantile_difficulty_binary",
)
ax.set_ylim(0, 3);
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:327: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond, "is_correct"])["rt"] /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:334: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond])["is_correct"]
plot_rt_hists(workshop_data,
by_participant = True,
split_by_column = "quantile_difficulty_binary",
inset_plot = "rt_mean")
/var/folders/gx/s43vynx550qbypcxm83fv56dzq4hgg/T/ipykernel_64548/900136031.py:142: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect. plt.tight_layout()
plot_rt_hists(workshop_data,
by_participant = True,
split_by_column = "quantile_difficulty_binary",
inset_plot = "choice_proportion")
/var/folders/gx/s43vynx550qbypcxm83fv56dzq4hgg/T/ipykernel_64548/900136031.py:142: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect. plt.tight_layout()
We see a similar pattern. Difficulty affects choice probability, however the effect on RT is less clear.
What parameter should difficulty map onto? Usually it maps onto the rate of evidence accumulation, which is the drift (v) parameter most SSMs.
We will move ahead and try this. To add specialized regression for v we can add another parameter dictionary to the list we pass to include.
Addressing difficulty¶
AngleHierModelV3 = hssm.HSSM(data = workshop_data,
model = "angle",
loglik_kind = "approx_differentiable",
global_formula = "y ~ (1|participant_id)",
include = [{"name": "a",
"formula": "a ~ (1 + C(costly_fail_condition)|participant_id)"},
{"name": "v",
"formula": "v ~ (1 + continuous_difficulty|participant_id)"},
],
noncentered = False,
)
Model initialized successfully.
AngleHierModelV3.graph()
try:
# Load pre-computed traces
AngleHierModelV3.restore_traces(traces = "scientific_workflow_hssm/idata/angle_hier_v3/traces.nc")
except:
# Sample posterior
angle_hier_idata = AngleHierModelV3.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
AngleHierModelV3.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
AngleHierModelV3.save_model(model_name = "angle_hier_v3",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
az.plot_posterior(AngleHierModelV3.traces,
var_names = ["v_continuous_difficulty|participant_id_mu"],
ref_val = 0,
kind = "hist",
ref_val_color = "red",
histtype = "step")
<Axes: title={'center': 'v_continuous_difficulty|participant_id_mu'}>
Looks like the effect on v is small (to the trained eye :)), but it is significant!
Let's check if we can account for the data pattern we missed previously.
ax = hssm.plotting.plot_quantile_probability(AngleHierModelV3,
cond="quantile_difficulty_binary",
)
ax.set_ylim(0, 3);
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:327: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond, "is_correct"])["rt"] /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:334: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond])["is_correct"]
# Posterior predictive
AngleHierModelV3.plot_predictive(step = True,
col = "quantile_difficulty_binary",
bins = np.linspace(-5,5, 50))
plt.tight_layout()
Success! This looks much better.
Quantitative Model Comparison¶
az.compare(
{
"DDM": BasicDDMModel.traces,
"DDM Hierarchical": DDMHierModel.traces,
"Angle Hierarchical": AngleHierModel.traces,
"Angle Hierarchical Cost": AngleHierModelV2.traces,
"Angle Hierarchical Cost/Diff": AngleHierModelV3.traces
}
)
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn( /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn(
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| Angle Hierarchical Cost/Diff | 0 | -4425.940827 | 69.185385 | 0.000000 | 9.876277e-01 | 70.554577 | 0.000000 | True | log |
| Angle Hierarchical Cost | 1 | -4470.826272 | 87.516854 | 44.885445 | 1.237269e-02 | 70.898643 | 9.559271 | False | log |
| Angle Hierarchical | 2 | -4736.150740 | 78.295641 | 310.209913 | 2.258851e-07 | 72.823473 | 22.959566 | False | log |
| DDM Hierarchical | 3 | -4954.407753 | 74.496783 | 528.466926 | 1.496612e-07 | 76.438410 | 27.593021 | True | log |
| DDM | 4 | -5645.531361 | 4.771162 | 1219.590534 | 0.000000e+00 | 74.728599 | 41.671983 | False | log |
Anything else?¶
At this point, we have a model that fits the data quite well.
We figured that a hierarchy significantly improves our fit, that the angle model dominates the basic ddm model for our data, and we incorporated effects based on
our experiment manipulations.
A natural next step is to check for patterns based on more generic properties of human choice data that we may be able to reason about.
Anything that comes to mind? Let's take another look at our dataset for some inspiration.
workshop_data
| response | rt | participant_id | trial | costly_fail_condition | continuous_difficulty | response_l1 | bin_difficulty | quantile_difficulty | quantile_difficulty_binary | response_l1_plotting | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0.556439 | 0 | 1 | 1 | -0.277337 | 0 | low | 0 | -1 | 1 |
| 1 | 1 | 0.741682 | 0 | 2 | 0 | -0.810919 | 1 | low | -1 | -1 | 1 |
| 2 | 1 | 0.461832 | 0 | 3 | 0 | -0.673330 | 1 | low | -1 | -1 | 1 |
| 3 | 1 | 0.626154 | 0 | 4 | 0 | 0.755445 | 1 | high | 1 | 1 | 1 |
| 4 | 1 | 0.651677 | 0 | 5 | 1 | 0.136755 | 1 | high | 0 | 1 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4995 | 1 | 1.039342 | 19 | 246 | 0 | -0.612223 | -1 | low | -1 | -1 | -1 |
| 4996 | 1 | 1.587827 | 19 | 247 | 0 | 0.732396 | 1 | high | 1 | 1 | 1 |
| 4997 | 1 | 0.668594 | 19 | 248 | 1 | -0.175321 | 1 | low | 0 | -1 | 1 |
| 4998 | 1 | 1.616471 | 19 | 249 | 0 | -0.630447 | 1 | low | -1 | -1 | 1 |
| 4999 | 1 | 1.051329 | 19 | 250 | 1 | 0.511197 | 1 | high | 1 | 1 | 1 |
5000 rows × 11 columns
At risk of stating the obvious,we have a column that went unused thus far: response_l1, the lagged response.
Maybe this hints at some level of stickiness in the choice behavior? How could we incorporate this?
Let us first investigate if there is indeed such a pattern in the data!
# Posterior predictive
AngleHierModelV3.plot_predictive(step = True,
col = 'response_l1_plotting',
bins = np.linspace(-5, 5, 50))
plt.tight_layout()
Indeed, it does seem like there is a bit of a pattern here, that we miss so far!
To incoporate choice stickiness, a reasonable candidate parameter is z, the a priori choice bias.
Maybe this parameter is affected by the last choice taken?
Let's try to incoporate this. We will
Addressing Stickyness¶
AngleHierModelV4 = hssm.HSSM(data = workshop_data,
model = "angle",
loglik_kind = "approx_differentiable",
global_formula = "y ~ (1|participant_id)",
include = [{"name": "a",
"formula": "a ~ (1 + C(costly_fail_condition)|participant_id)"},
{"name": "v",
"formula": "v ~ (1 + continuous_difficulty|participant_id)"},
{"name": "z",
"formula": "z ~ (1 + response_l1|participant_id)"},
],
noncentered = False,
)
Model initialized successfully.
AngleHierModelV4.graph()
try:
# Load pre-computed traces
AngleHierModelV4.restore_traces(traces = "scientific_workflow_hssm/idata/angle_hier_v4/traces.nc")
except:
# Sample posterior
angle_hier_idata = AngleHierModelV4.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
AngleHierModelV4.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
AngleHierModelV4.save_model(model_name = "angle_hier_v4",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
az.plot_trace(AngleHierModelV4.traces,
divergences = None);
plt.tight_layout()
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/plots/traceplot.py:223: UserWarning: rcParams['plot.max_subplots'] (20) is smaller than the number of variables to plot (24), generating only 20 plots warnings.warn(
** Note **:
We can see some rather interesting artifacts in the chains above. Around samples 300-375 it looks like our solid-blue chain got quite stuck. This indicates some problems with the posterior geometry for this model.
One diagnostic that can be helpful here whether or not we observe a lot of divergences during sampling.
Let's take a look below (notice, we change the diveregences argument from None to it's default)
az.plot_trace(AngleHierModelV4.traces,
divergences = 'auto');
plt.tight_layout()
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/plots/traceplot.py:223: UserWarning: rcParams['plot.max_subplots'] (20) is smaller than the number of variables to plot (24), generating only 20 plots warnings.warn(
Indeed, we observe a fee divergences here... as rigorous scientists, we should now try to get to the bottom of this phenomenon (it happens often if one tries hierarchical models naively on real experimental data). In the context of this tutorial, we will let it slide however. It would warrant a longer detour.
Let's move on and focus on whether or not we actually identify a significant choice stickyness effect with our analysis:
az.plot_posterior(AngleHierModelV4.traces,
var_names = ["z_response_l1|participant_id_mu"],
ref_val = 0,
kind = "hist",
ref_val_color = "red",
histtype = "step")
<Axes: title={'center': 'z_response_l1|participant_id_mu'}>
We observe a significant effect on the z parameter, in fact a mean effect of 0.073 insinuate a fairly big effect of choice stickyness.
In direct comparison, we might expect this effect to overall have a larger impact on our model fit than the effect of difficulty on v, which we investigated in the
previous section.
# Posterior predictive
AngleHierModelV4.plot_predictive(step = True,
col = 'response_l1_plotting',
bins = np.linspace(-5, 5, 50))
plt.tight_layout()
Quantitative Model Comparison¶
az.compare(
{"DDM": BasicDDMModel.traces,
"DDM Hierarchical": DDMHierModel.traces,
"Angle Hierarchical": AngleHierModel.traces,
"Angle Hierarchical Cost": AngleHierModelV2.traces,
"Angle Hierarchical Cost/Diff": AngleHierModelV3.traces,
"Angle Hierarchical Cost/Diff/Sticky": AngleHierModelV4.traces}
)
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn( /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn(
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| Angle Hierarchical Cost/Diff/Sticky | 0 | -4327.769237 | 103.455670 | 0.000000 | 0.918195 | 71.004980 | 0.000000 | False | log |
| Angle Hierarchical Cost/Diff | 1 | -4425.940827 | 69.185385 | 98.171590 | 0.081805 | 70.554577 | 15.685768 | True | log |
| Angle Hierarchical Cost | 2 | -4470.826272 | 87.516854 | 143.057036 | 0.000000 | 70.898643 | 17.113407 | False | log |
| Angle Hierarchical | 3 | -4736.150740 | 78.295641 | 408.381504 | 0.000000 | 72.823473 | 26.073614 | False | log |
| DDM Hierarchical | 4 | -4954.407753 | 74.496783 | 626.638516 | 0.000000 | 76.438410 | 29.304281 | True | log |
| DDM | 5 | -5645.531361 | 4.771162 | 1317.762125 | 0.000000 | 74.728599 | 43.174373 | False | log |
And indeed, the drop inelpd_loo is even more substantial, than the improvement generated by incorporating the difficulty effect.
Taking Stock¶
ax = hssm.plotting.plot_quantile_probability(AngleHierModelV4,
cond="response_l1_plotting",
)
ax.set_ylim(0, 3);
ax = hssm.plotting.plot_quantile_probability(AngleHierModelV4,
cond="costly_fail_condition",
)
ax.set_ylim(0, 3);
ax = hssm.plotting.plot_quantile_probability(AngleHierModelV4,
cond="quantile_difficulty_binary",
)
ax.set_ylim(0, 3);
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:327: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond, "is_correct"])["rt"] /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/src/hssm/plotting/utils.py:334: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning. df.groupby(["observed", "chain", "draw", cond])["is_correct"]
Sanity Check, was the hierarchy really necessary¶
AngleModelV5 = hssm.HSSM(data = workshop_data,
model = "angle",
loglik_kind = "approx_differentiable",
global_formula = "y ~ 1",
include = [{"name": "a",
"formula": "a ~ 1 + C(costly_fail_condition)"},
{"name": "v",
"formula": "v ~ 1 + continuous_difficulty"},
{"name": "z",
"formula": "z ~ 1 + response_l1"},
],
noncentered = False,
)
Model initialized successfully.
try:
# Load pre-computed traces
AngleModelV5.restore_traces(traces = "scientific_workflow_hssm/idata/angle_v5/traces.nc")
except:
# Sample posterior
angle_hier_idata = AngleModelV5.sample(chains = 2,
sampler = "nuts_numpyro",
tune = 500,
draws = 500,
)
# Sample posterior predictive
AngleModelV5.sample_posterior_predictive(draws = 200,
safe_mode = True)
# Save Model
AngleModelV5.save_model(model_name = "angle_v5",
allow_absolute_base_path = True,
base_path = "scientific_workflow_hssm/idata/",
save_idata_only = True)
az.compare(
{
"DDM": BasicDDMModel.traces,
"DDM Hierarchical": DDMHierModel.traces,
"Angle Hierarchical": AngleHierModel.traces,
"Angle Hierarchical Cost": AngleHierModelV2.traces,
"Angle Hierarchical Cost/Diff": AngleHierModelV3.traces,
"Angle Hierarchical Cost/Diff/Sticky": AngleHierModelV4.traces,
"Angle Cost/Diff/Sticky": AngleModelV5.traces
}
)
/Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn( /Users/afengler/Library/CloudStorage/OneDrive-Personal/proj_hssm/HSSM/.venv/lib/python3.11/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.67 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations. warnings.warn(
| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| Angle Hierarchical Cost/Diff/Sticky | 0 | -4327.769237 | 103.455670 | 0.000000 | 0.918195 | 71.004980 | 0.000000 | False | log |
| Angle Hierarchical Cost/Diff | 1 | -4425.940827 | 69.185385 | 98.171590 | 0.081805 | 70.554577 | 15.685768 | True | log |
| Angle Hierarchical Cost | 2 | -4470.826272 | 87.516854 | 143.057036 | 0.000000 | 70.898643 | 17.113407 | False | log |
| Angle Hierarchical | 3 | -4736.150740 | 78.295641 | 408.381504 | 0.000000 | 72.823473 | 26.073614 | False | log |
| DDM Hierarchical | 4 | -4954.407753 | 74.496783 | 626.638516 | 0.000000 | 76.438410 | 29.304281 | True | log |
| Angle Cost/Diff/Sticky | 5 | -5001.726336 | 8.676713 | 673.957099 | 0.000000 | 70.471189 | 33.148987 | False | log |
| DDM | 6 | -5645.531361 | 4.771162 | 1317.762125 | 0.000000 | 74.728599 | 43.174373 | False | log |
The End:¶
So far so good, we completed a rather comprehensive model exploration and we generated quite a few insights! We could obviously go on and try more and more complex models and maybe there is more to find out here... we leave this up to you and hope that HSSM will continue to help you along the way :).
Pointers to advanced Topics¶
We are scratching only the surface of what cann be done with HSSM, let alone the broader eco-system supporting simulation based inference (SBI).
Check out our simulator package, ssm-simulators as well as our our little neural network library for training LANs, lanfactory.
Exciting work is being done (more on this in the next tutorial) on connecting to other packages in the wider eco-system, such as BayesFlow as well as the sbi package.
Here is a taste of advanced topics with links to corresponding tutorials:
- Variational Inference with HSSM
- Build PyMC models with HSSM random variables
- Connect compiled models to third party MCMC libraries
- Construct custom models from simulators and contributed likelihoods
- Using link functions to transform parameters
you will find this and a lot more information in the official documentation